Summary: The third pipeline produces a machine learning model that you can use to predict the disease status of new samples (healthy/disease). With this modelyou can create a new biomarker for your disease based on sRNA sequence count data.
This tutorial explains how to submit count files (output by Oasis' sRNA Detection module) to the Oasis Classification module. The Classification module is the third analysis module of Oasis and it performs a binary classification on the count files by applying a Random Forest classifier. This step in the Oasis workflow provides an exploratory analysis of the data, gives classifier's performance measures and a list of the most relevant features (i.e. sRNAs) in classifying the different samples into their equivalent groups. Guidelines on how to interpret the results of the Classification detection module can be found in Oasis' Classification Output Tutorial.
Like any other Oasis job, you need to fill in the fields for the E-Mail address where status updates and results should be mailed to. You also have to pick an Experiment Name that will later allow you to remember what the Oasis job was about when you submitted it. The Reference Genome should correspond to the species from which the samples have been taken.

For the control group and the treatment group(s) you need to upload sRNA count files from your computer.
You probably have obtained these sRNA count files from downloading your results from the sRNA detection module.
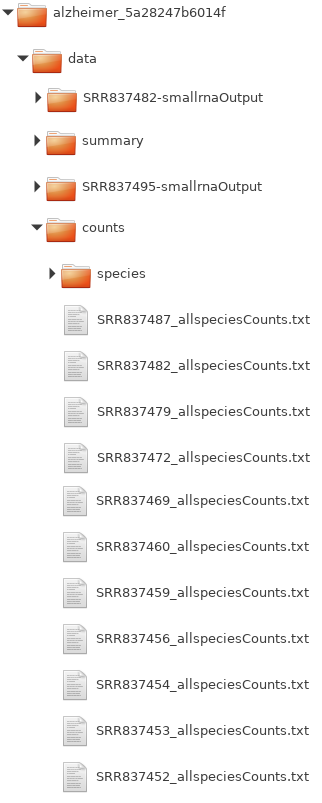
If you look at the sRNA detection module results, the folder structure is as
depicted in figure : You need to acess the
data/counts/ subfolder and then pick those
_allspeciesCounts.txt files that correspond to either the control group
or one of the treatment groups. Note that you can select multiple files for each
upload field at once. This way you can bundle all control count files and all
treatment count files (for each group).
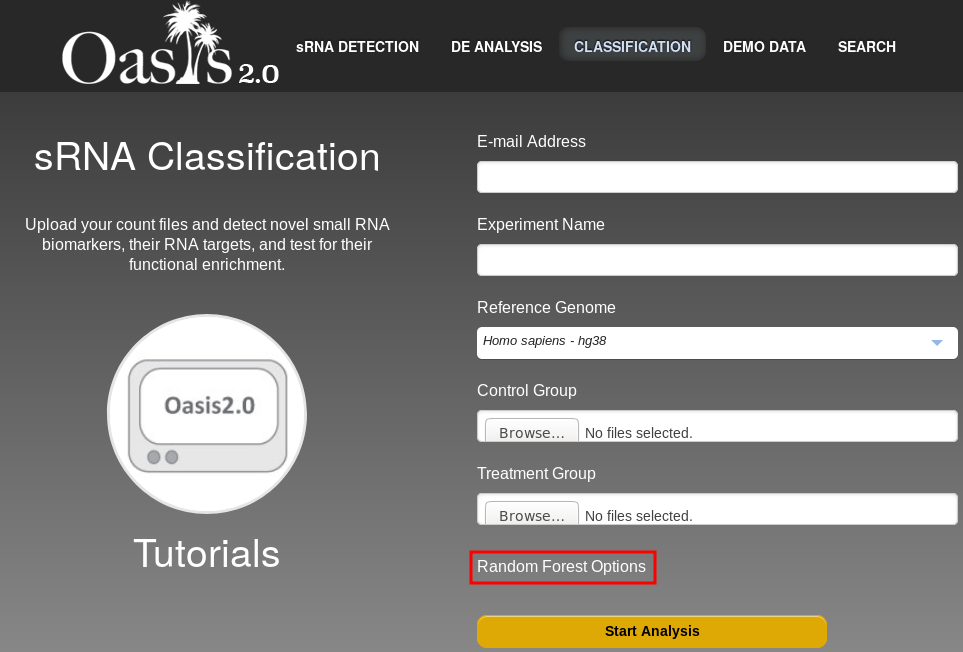
If you click on Random Forest Options (marked in a red box in figure ) you will have access to two more form fields:
Capece, V., Garcia Vizcaino, J. C., Vidal, R., Rahman, R.-U., Pena Centeno, T., Shomroni, O., ... Bonn, S. (2015).
Oasis: online analysis of small RNA deep sequencing data. Bioinformatics, 31 (13), 2205-2207.
http://doi.org/10.1093/bioinformatics/btv113